Hi friends!
My name is Annabel Large (国晓瑛), I use she/her pronouns, and I’m a 3rd year PhD candidate advised by Ian Holmes. I’m housed in the Bioengineering graduate group at the University of California (UC), Berkeley and UC San Francisco. I’m also pursuing a Designated Emphasis in Computational and Genomic Biology through UC Berkeley’s Center for Computational Biology.
I’m working on building statistical and machine learning models to better characterize and understand the engineering of Adeno-associated virus (AAV) capsids. Specifically, with all the newest tools and toys from Nanopore informatics and protein language modeling, can we discover new parts of AAV functional landscape? (see my Up-Goer Five summary attempt below!)
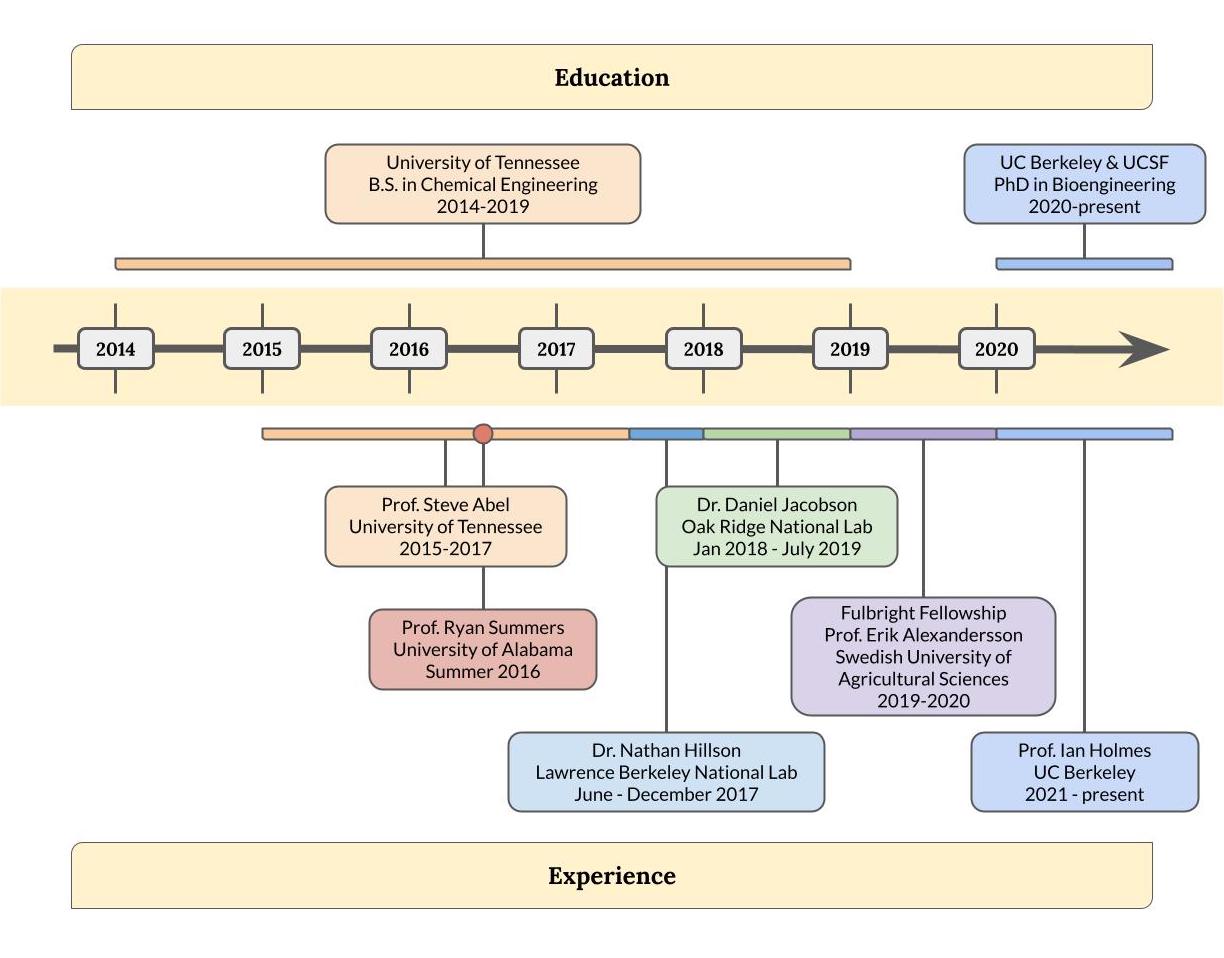
Graphical CV
Up-Goer Five Research Statement
‘Adeno_associated_virus’ can carry things needed to fix a broken cell, but only if ‘AAV’ is carefully made. Making better ‘AAV’ is hard because there are many different orders ‘AAV’ parts can be in. I want to use computers to understand how to make the best ‘AAV’ for a given job. Even deeper, I hope to learn why these orders work. Why does a part need to be in that position? What parts could make the whole ‘AAV’ break? Computers can help us learn these controls! :)